Structure rotation
-
Hello!
I am struggling to understand the way in which CRYSTAL rotates the atoms. Here is my structure:
reference.f34
I want to rotate all atoms so that Mn remains in place, and oxygens NN 21&23 move to the ac (xz) plane. This, in essence, is a rotation around the axis 10Mn-12P, the result of which should be that
Y coordinate of Mn = Y(O21) = Y(O23). The angle that should work is +-24.7159 degrees, depending on the direction that CRYSTAL uses.
So, I triedATOMROT 0 # include all atoms 999 1 # no translation & rotation around a specified axis 10 12 # atom labels that define rotation axis +/-24.7159 # angle (degrees)To the effect of
ALL THE 36 ATOMS IN THE REFERENCE CELL ARE MANIPULATED SELECTED ATOMS ARE ROTATED AROUND THE AXIS DEFINED BY TWO ATOMS: 10 12 (DIRECTION COSINES 0.000 0.000 -1.000) BY AN ANGLE: 24.72 ATOM 10 AT.N. 25 COORDINATES 4.46889 0.00000 2.24246 ATOM 21 AT.N. 8 COORDINATES -2.74300 2.67751 3.17915 ATOM 23 AT.N. 8 COORDINATES 3.56177 1.05955 3.17915and
ALL THE 36 ATOMS IN THE REFERENCE CELL ARE MANIPULATED SELECTED ATOMS ARE ROTATED AROUND THE AXIS DEFINED BY TWO ATOMS: 10 12 (DIRECTION COSINES 0.000 0.000 -1.000) BY AN ANGLE: -24.72 ATOM 10 AT.N. 25 COORDINATES 4.46889 0.00000 2.24246 ATOM 21 AT.N. 8 COORDINATES -2.25530 -3.73706 3.17915 ATOM 23 AT.N. 8 COORDINATES 3.07407 0.00001 3.17915So the second method maybe worked? Except that I have been unable to visualize/confirm it using VESTA, because the output of edited geometry section looks the same as the input*.
So, my questions are:
- how to validate the result of rotation?
- when using ROTCRY+MATROT, where is the origin of rotation?
- which method of rotation I should use in this situation?
*VESTA, of course, does not open .f34 files, so my usual workflow is to convert CRYSTAL output to a VASP-POSCAR format:
Structure description line 1.0 #scaling factor [DIRECT LATTICE VECTORS CARTESIAN COMPONENTS (ANGSTROM)] List-of-elements Amounts-of-each-element Direct or Cartesian # coordinate format [list of coordinates in the selected format] -
What I have realised is that I may not need to perform an actual rotation. Quote from the manual:
A rotation of the eigenvectors can be
obtained in properties by entering the keyword ROTREF, allowing AO projected Density
of States or Population Analysis orienting the cartesian frame along the principal axes of the
octahedron.I was hoping that defining a plane with three atoms that include the Mn-P axis and one of the two O atoms would be sufficient
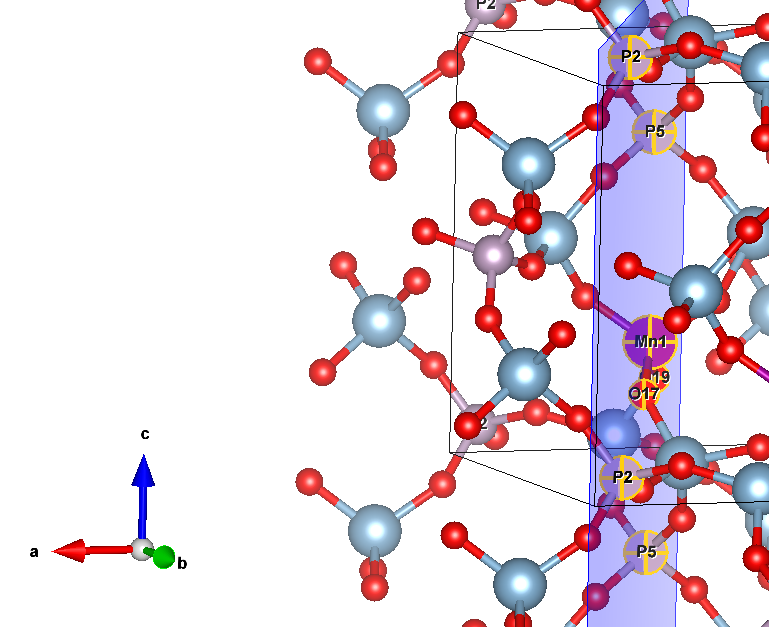
For instance,ROTREF ATOMS 10 # this is Manganese 0 0 0 # I also tried this with different cell indices 12 # P5 0 0 0 8 # P2 0 0 0 NEWK 8 8 1 0 DOSSOr , when that resulted in an error immediately after k-point generation, I thought that maybe changing k-points will help, but both
NEWK 1 1 1 0and
NEWK 11 11 1 0end up in
181-C( 6 3 5) 182-C( 7 3 5) 183-C( 4 4 5) 184-C( 5 4 5) 185-C( 6 4 5) 186-C( 5 5 5) -------------------------------------------------------------------------- MPI_ABORT was invoked on rank 19 in communicator MPI_COMM_WORLD with errorcode 1. NOTE: invoking MPI_ABORT causes Open MPI to kill all MPI processes. You may or may not see output from other processes, depending on exactly when Open MPI kills them. -------------------------------------------------------------------------- [lasc110:51218] PMIX ERROR: UNREACHABLE in file server/pmix_server.c at line 2198 forrtl: error (78): process killed (SIGTERM) Image PC Routine Line Source Pproperties 00000000061B7B8B for__signal_handl Unknown Unknown libpthread-2.17.s 00007FE293190630 Unknown Unknown Unknown Pproperties 000000000141ADBC Unknown Unknown Unknown Pproperties 0000000000548C2C Unknown Unknown Unknown Pproperties 0000000000543D3B Unknown Unknown Unknown Pproperties 000000000040F5DC Unknown Unknown Unknown Pproperties 000000000040DB92 Unknown Unknown Unknown Pproperties 000000000040D9A2 Unknown Unknown Unknown libc-2.17.so 00007FE292DD5555 __libc_start_main Unknown Unknown Pproperties 000000000040D8AA Unknown Unknown Unknown -
Poking further, since the last error is that of MPI and not of CRYSTAL, I launched the same input with NEWK 8 8 as a single-core process (with Pproperties), and this time the error was different:
85-C( 3 2 4) 86-C( 4 2 4) 87-C( 5 2 4) 88-C( 3 3 4) 89-C( 4 3 4) 90-R( 4 4 4) ERROR **** PROJVR **** NULL COMPONENT 0.222045E-15 0.100000E-07for
ROTREF ATOMS 10 # this is Manganese 0 0 0 # I also tried this with different cell indices 12 # P5 0 0 0 8 # P2 0 0 0and then
85-C( 3 2 4) 86-C( 4 2 4) 87-C( 5 2 4) 88-C( 3 3 4) 89-C( 4 3 4) 90-R( 4 4 4) ERROR **** RHOLSK **** BASIS SET LINEARLY DEPENDENTfor the following input:
ROTREF ATOMS 10 1 0 0 12 1 0 1 8 1 1 0As well as for other indices
