Hello!
I am struggling to understand the way in which CRYSTAL rotates the atoms. Here is my structure:
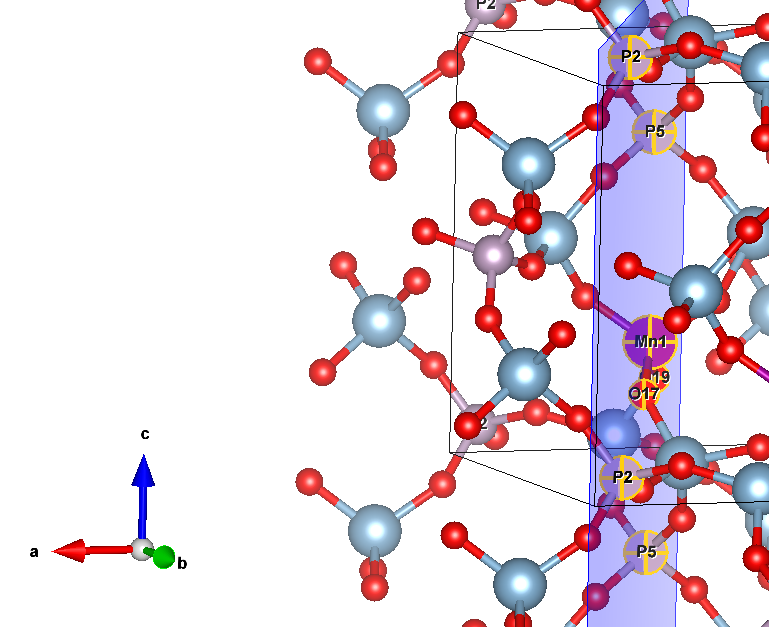
reference.f34
I want to rotate all atoms so that Mn remains in place, and oxygens NN 21&23 move to the ac (xz) plane. This, in essence, is a rotation around the axis 10Mn-12P, the result of which should be that
Y coordinate of Mn = Y(O21) = Y(O23). The angle that should work is +-24.7159 degrees, depending on the direction that CRYSTAL uses.
So, I tried
ATOMROT
0 # include all atoms
999 1 # no translation & rotation around a specified axis
10 12 # atom labels that define rotation axis
+/-24.7159 # angle (degrees)
To the effect of
ALL THE 36 ATOMS IN THE REFERENCE CELL ARE MANIPULATED
SELECTED ATOMS ARE ROTATED AROUND THE AXIS DEFINED BY TWO ATOMS: 10 12
(DIRECTION COSINES 0.000 0.000 -1.000) BY AN ANGLE: 24.72
ATOM 10 AT.N. 25 COORDINATES 4.46889 0.00000 2.24246
ATOM 21 AT.N. 8 COORDINATES -2.74300 2.67751 3.17915
ATOM 23 AT.N. 8 COORDINATES 3.56177 1.05955 3.17915
and
ALL THE 36 ATOMS IN THE REFERENCE CELL ARE MANIPULATED
SELECTED ATOMS ARE ROTATED AROUND THE AXIS DEFINED BY TWO ATOMS: 10 12
(DIRECTION COSINES 0.000 0.000 -1.000) BY AN ANGLE: -24.72
ATOM 10 AT.N. 25 COORDINATES 4.46889 0.00000 2.24246
ATOM 21 AT.N. 8 COORDINATES -2.25530 -3.73706 3.17915
ATOM 23 AT.N. 8 COORDINATES 3.07407 0.00001 3.17915
So the second method maybe worked? Except that I have been unable to visualize/confirm it using VESTA, because the output of edited geometry section looks the same as the input*.
So, my questions are:
- how to validate the result of rotation?
- when using ROTCRY+MATROT, where is the origin of rotation?
- which method of rotation I should use in this situation?
*VESTA, of course, does not open .f34 files, so my usual workflow is to convert CRYSTAL output to a VASP-POSCAR format:
Structure description line
1.0 #scaling factor
[DIRECT LATTICE VECTORS CARTESIAN COMPONENTS (ANGSTROM)]
List-of-elements
Amounts-of-each-element
Direct or Cartesian # coordinate format
[list of coordinates in the selected format]