Davide, a follow up question. Will that not affect the resulting thermodynamics? I mean translational or rotational (I can never remember) thermodynamics is dependent on symmetry and I am calculating thermodynamic quantities here
Jonas Baltrusaitis
Posts
-
D infinity h symmetry -
D infinity h symmetryFor the isolated molecule such as CO2 which is D infinity h, what is the symmetry group? I could not find this one in the manual
-
extract asymmetric fragmentvery grateful
-
SCANMODE io error Read_int_1dSo it turns out having fort.13 and fort.20 is not optional for restart. Now I still need help. I optimizing this 600 atom cluster with Ih and can see that it is not minimum, there are negatives - this is why I am scanning. The scan does not return a reasonable local minimum. I only scanned negative branch but I assume they are symmetrical.
I would appreciate any helphttps://www.dropbox.com/t/5jgMXmCEPNLGHpAY
PS. I would also like to ask developers to allow uploading compressed files to this forum
SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS SCAN ALONG NORMAL MODES STARTING POINT: -20 ENDING POINT: 20 STEP: 0.20000 (THE STEP IS GIVEN AS TIMES OF THE CLASSICAL AMPLITUDE AT THE QUANTUM GROUND STATE ENERGY. THE MAX ATOMIC DISPLACEMENT IN THE STEP IS GIVEN IN BOHR WITHIN SQUARE BRACKETS) MODE(CM-1) DISPLAC TOTAL ENE(DFT)(AU) CLASSICAL HARM ENE(AU) NCYC DE [MAX DISP -NATOM-] 1( -11.5) [ 0.016 - 512-] -4.0000 -0.2201822589948E+05 -0.2201822692117E+05 15 -0.7E-09 -3.8000 -0.2201822596504E+05 -0.2201822688013E+05 15 -0.5E-09 -3.6000 -0.2201822602580E+05 -0.2201822684119E+05 15 -0.5E-09 -3.4000 -0.2201822608201E+05 -0.2201822680436E+05 16 0.3E-10 -3.2000 -0.2201822613398E+05 -0.2201822676964E+05 14 0.1E-08 -3.0000 -0.2201822618174E+05 -0.2201822673702E+05 17 0.1E-10 -2.8000 -0.2201822622563E+05 -0.2201822670650E+05 17 -0.7E-10 -2.6000 -0.2201822626589E+05 -0.2201822667809E+05 18 0.2E-10 -2.4000 -0.2201822630238E+05 -0.2201822665178E+05 16 -0.9E-10 -2.2000 -0.2201822633530E+05 -0.2201822662758E+05 17 0.0E+00 -2.0000 -0.2201822636488E+05 -0.2201822660548E+05 16 0.4E-10 -1.8000 -0.2201822639128E+05 -0.2201822658549E+05 16 -0.4E-10 -1.6000 -0.2201822641463E+05 -0.2201822656760E+05 15 -0.4E-10 -1.4000 -0.2201822643453E+05 -0.2201822655181E+05 17 0.4E-10 -1.2000 -0.2201822645245E+05 -0.2201822653813E+05 14 0.1E-10 -1.0000 -0.2201822646714E+05 -0.2201822652656E+05 15 -0.5E-10 -0.8000 -0.2201822647907E+05 -0.2201822651709E+05 14 -0.4E-10 -0.6000 -0.2201822648831E+05 -0.2201822650972E+05 15 -0.4E-10 -0.4000 -0.2201822649498E+05 -0.2201822650446E+05 14 0.3E-10 -0.2000 -0.2201822649893E+05 -0.2201822650130E+05 12 -0.6E-10 0.0000 -0.2201822650025E+05 -0.2201822650025E+05 CENTRAL POINT -
SCANMODE io error Read_int_1dI lost my patience... I normally do OPT+FREQ, nobody's on this forum (even developers) favorite method... So I just took my optimization, did frequencies, found fort.13 file, included and scanmode proceeded. I will update if anything else happens.
-
SCANMODE io error Read_int_1dOh, no, Aleks. It did not even start, e.g. it was setting up the scan, about to start the first step and stopped. The lines are the end of my output file, no optimization steps, no SCFOUT.LOG
-
SCANMODE io error Read_int_1dDear all, I am getting the IO error when scanning modes and no idea why. I copied HESSFREQ.DAT and FREQINFO.DAT files into the scan directory. I would appreciate your help
Unfortunately, it is a large system and I can't upload the files here, I will move them via Dropbox, see the link below
https://www.dropbox.com/t/uVxYAZdbhCFpM79K
SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS SCAN ALONG NORMAL MODES STARTING POINT: -10 ENDING POINT: 10 STEP: 0.40000 (THE STEP IS GIVEN AS TIMES OF THE CLASSICAL AMPLITUDE AT THE QUANTUM GROUND STATE ENERGY. THE MAX ATOMIC DISPLACEMENT IN THE STEP IS GIVEN IN BOHR WITHIN SQUARE BRACKETS) MODE(CM-1) DISPLAC TOTAL ENE(DFT)(AU) CLASSICAL HARM ENE(AU) NCYC DE [MAX DISP -NATOM-] 1( -8.5) [ 0.030 - 390-] io error Read_int_1d 300 -1 Abort(1) on node 0 (rank 0 in comm 0): application called MPI_Abort(MPI_COMM_WORLD, 1) - process 0 -
extract asymmetric fragmentI have this 600 atom Ih symmetry molecular cage. I am seeking help extracting the asymmetric unit from it to enter into CRYSTAL so I can use Ih symmetry. If anybody could help me with it, I would appreciate it.
JB
-
SCANMODE problemHi Aleks, I tried but ended up just using LDREMO keyword
Another superstrange thing (that of course was not happening yesterday), that I can't seem to specify initial and final step anymore properly, it divides it by 10... Bizzare. I now have to multiply initial step by 10 (-1.6 start I want I need to specify as 16!) so it startes at -1.6...My input is:
EXTERNAL FREQCALC RESTART SCANMODE 1 -16 0 0.1 1 END ENDSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS SCAN ALONG NORMAL MODES STARTING POINT: -16 ENDING POINT: 0 STEP: 0.10000 (THE STEP IS GIVEN AS TIMES OF THE CLASSICAL AMPLITUDE AT THE QUANTUM GROUND STATE ENERGY. THE MAX ATOMIC DISPLACEMENT IN THE STEP IS GIVEN IN BOHR WITHIN SQUARE BRACKETS) MODE(CM-1) DISPLAC TOTAL ENE(DFT)(AU) CLASSICAL HARM ENE(AU) NCYC DE [MAX DISP -NATOM-] 1( -62.7) [ 0.028 - 3-] -1.6000 -0.1610014638959E+04 -0.1610015023766E+04 18 -0.1E-07 -1.5000 -0.1610014685887E+04 -0.1610014979469E+04 29 0.5E-06 -1.4000 -0.1610014719434E+04 -0.1610014938029E+04 13 -0.9E-07 -1.3000 -0.1610014740251E+04 -0.1610014899447E+04 13 -0.2E-06 -1.2000 -0.1610014750671E+04 -0.1610014863723E+04 21 -0.9E-08 -1.1000 -0.1610014754239E+04 -0.1610014830856E+04 15 -0.4E-06 -1.0000 -0.1610014749993E+04 -0.1610014800848E+04 28 0.1E-07 -0.9000 -0.1610014740870E+04 -0.1610014773698E+04 23 0.1E-05 -
SCANMODE problemOh, I see... somewhere among the files I found this. It is unusual since everything converged optimized/converged otherwise. Now wonder I can't restart, it is not restart problem. Why would i become linearly dependent in SCANMODE but not before?
ERROR **** CHOLSK **** BASIS SET LINEARLY DEPENDENT
-
SCANMODE problemNo idea... I just have no idea... I ran a complete run and all of a sudden the same abort. So frustrating since I do not understand the error
SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS SCAN ALONG NORMAL MODES STARTING POINT: -1 ENDING POINT: 0 STEP: 20.00000 (THE STEP IS GIVEN AS TIMES OF THE CLASSICAL AMPLITUDE AT THE QUANTUM GROUND STATE ENERGY. THE MAX ATOMIC DISPLACEMENT IN THE STEP IS GIVEN IN BOHR WITHIN SQUARE BRACKETS) MODE(CM-1) DISPLAC TOTAL ENE(DFT)(AU) CLASSICAL HARM ENE(AU) NCYC DE [MAX DISP -NATOM-] 1( -62.7) [ 5.606 - 3-] Abort(1) on node 31 (rank 31 in comm 0): application called MPI_Abort(MPI_COMM_WORLD, 1) - process 31 -
SCANMODE problemfort.f34 INPUT.d12 job.out Further on this problem - I managed to run this without restart - I am notoriously bad restarting. Tryiong to get rid of this small negative frequenyc. I do not see from the scan any problems, e.g. any local minimum I could restart calculation from. It just decreases monotonically...
MODE(CM-1) DISPLAC TOTAL ENE(DFT)(AU) CLASSICAL HARM ENE(AU) NCYC DE [MAX DISP -NATOM-] 1( -62.1) [ 0.113 - 3-] -4.0000 -0.1610002871486E+04 -0.1610016919330E+04 43 -0.9E-10 -3.6000 -0.1610006913204E+04 -0.1610016489435E+04 32 -0.4E-10 -3.2000 -0.1610009918764E+04 -0.1610016104792E+04 34 0.2E-11 -2.8000 -0.1610012035843E+04 -0.1610015765400E+04 37 -0.8E-11 -2.4000 -0.1610013425826E+04 -0.1610015471261E+04 39 0.2E-08 -2.0000 -0.1610014243529E+04 -0.1610015222375E+04 121 0.4E-09 -1.6000 -0.1610014634134E+04 -0.1610015018740E+04 39 -0.5E-12 -1.2000 -0.1610014749019E+04 -0.1610014860357E+04 78 0.4E-10 -0.8000 -0.1610014729596E+04 -0.1610014747227E+04 54 -0.1E-09 -0.4000 -0.1610014679529E+04 -0.1610014679349E+04 36 0.1E-11 0.0000 -0.1610014656723E+04 -0.1610014656723E+04 CENTRAL POINT -
SCANMODE problemHi all, trying to eliminate a negative frequency via SCANMODE. Job aborts with no particular message before scanning. I have FREQINFO file for the restart
FREQINFO.DAT INPUT.d12 input.f34 input.outEXTERNAL FREQCALC RESTART SCANMODE 1 -10 10 0.4 1 END ENDSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS SCAN ALONG NORMAL MODES STARTING POINT: -10 ENDING POINT: 10 STEP: 0.40000 (THE STEP IS GIVEN AS TIMES OF THE CLASSICAL AMPLITUDE AT THE QUANTUM GROUND STATE ENERGY. THE MAX ATOMIC DISPLACEMENT IN THE STEP IS GIVEN IN BOHR WITHIN SQUARE BRACKETS) MODE(CM-1) DISPLAC TOTAL ENE(DFT)(AU) CLASSICAL HARM ENE(AU) NCYC DE [MAX DISP -NATOM-] 1( -62.1) [ 0.113 - 3-] -
fractional coordinate entryHi Giu, it worked!
-
fractional coordinate entryHi all, I am not getting the right geometry inputs for 1/3 and 2/3 positions. For example, for the structure below, I am trying to enter for the group 164:
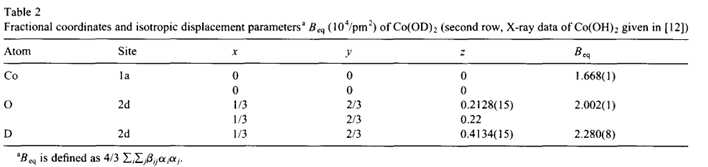
CRYSTAL
0 0 0
164
3.186 4.653
3
27 0.0 0.0 0.0
1 0.333 0.6666 0.4134
8 0.3333 0.666 0.2128and it ends up being incorrect stoichiometry, which tells me these fractional positions were not parsed correctly.
CRYSTAL CALCULATION
(INPUT ACCORDING TO THE INTERNATIONAL TABLES FOR X-RAY CRYSTALLOGRAPHY)
CRYSTAL FAMILY : HEXAGONAL
CRYSTAL CLASS (GROTH - 1921) : DITRIGONAL SCALENOHEDRALSPACE GROUP (CENTROSYMMETRIC) : P -3 M 1
LATTICE PARAMETERS (ANGSTROMS AND DEGREES) - CONVENTIONAL CELL
A B C ALPHA BETA GAMMA
3.18600 3.18600 4.65300 90.00000 90.00000 120.00000NUMBER OF IRREDUCIBLE ATOMS IN THE CONVENTIONAL CELL: 3
INPUT COORDINATES
ATOM AT. N. COORDINATES
1 27 0.000000000000E+00 0.000000000000E+00 0.000000000000E+00
2 1 3.330000000000E-01 6.666000000000E-01 4.134000000000E-01
3 8 3.333000000000E-01 6.660000000000E-01 2.128000000000E-01
<< INFORMATION >>: FROM NOW ON, ALL COORDINATES REFER TO THE PRIMITIVE CELL
LATTICE PARAMETERS (ANGSTROMS AND DEGREES) - PRIMITIVE CELL
A B C ALPHA BETA GAMMA VOLUME
3.18600 3.18600 4.65300 90.00000 90.00000 120.00000 40.903006COORDINATES OF THE EQUIVALENT ATOMS (FRACTIONAL UNITS)
N. ATOM EQUIV AT. N. X Y Z
1 1 1 27 CO 0.00000000000E+00 0.00000000000E+00 0.00000000000E+00
2 2 1 1 H 3.33000000000E-01 -3.33400000000E-01 4.13400000000E-01
3 2 2 1 H 3.33400000000E-01 -3.33600000000E-01 4.13400000000E-01
4 2 3 1 H 3.33600000000E-01 -3.33000000000E-01 4.13400000000E-01
5 2 4 1 H -3.33600000000E-01 3.33400000000E-01 -4.13400000000E-01
6 2 5 1 H -3.33000000000E-01 3.33600000000E-01 -4.13400000000E-01
7 2 6 1 H -3.33400000000E-01 3.33000000000E-01 -4.13400000000E-01
8 2 7 1 H -3.33000000000E-01 3.33400000000E-01 -4.13400000000E-01
9 2 8 1 H -3.33400000000E-01 3.33600000000E-01 -4.13400000000E-01
10 2 9 1 H -3.33600000000E-01 3.33000000000E-01 -4.13400000000E-01
11 2 10 1 H 3.33600000000E-01 -3.33400000000E-01 4.13400000000E-01
12 2 11 1 H 3.33000000000E-01 -3.33600000000E-01 4.13400000000E-01
13 2 12 1 H 3.33400000000E-01 -3.33000000000E-01 4.13400000000E-0114 3 1 8 O 3.33300000000E-01 -3.34000000000E-01 2.12800000000E-01
15 3 2 8 O 3.34000000000E-01 -3.32700000000E-01 2.12800000000E-01
16 3 3 8 O 3.32700000000E-01 -3.33300000000E-01 2.12800000000E-01
17 3 4 8 O -3.32700000000E-01 3.34000000000E-01 -2.12800000000E-01
18 3 5 8 O -3.33300000000E-01 3.32700000000E-01 -2.12800000000E-01
19 3 6 8 O -3.34000000000E-01 3.33300000000E-01 -2.12800000000E-01
20 3 7 8 O -3.33300000000E-01 3.34000000000E-01 -2.12800000000E-01
21 3 8 8 O -3.34000000000E-01 3.32700000000E-01 -2.12800000000E-01
22 3 9 8 O -3.32700000000E-01 3.33300000000E-01 -2.12800000000E-01
23 3 10 8 O 3.32700000000E-01 -3.34000000000E-01 2.12800000000E-01
24 3 11 8 O 3.33300000000E-01 -3.32700000000E-01 2.12800000000E-01
25 3 12 8 O 3.34000000000E-01 -3.33300000000E-01 2.12800000000E-01 -
Is there a software to create DOS/BAND input?wonderful, thank you both
-
Is there a software to create DOS/BAND input?thank you, this is very useful for me
-
TREATMENT OF DISORDERED SYSTEMS / SOLID SOLUTIONS - perovskite structureHi, we synthesized a complex material something along the lines of LaFex Niy Coz Cuu Mnv O3 where x+y+z+u+n=1. I know it is not trivial, but I would like to attempt using configuration analysis in TREATMENT OF DISORDERED SYSTEMS / SOLID SOLUTIONS module to generate possible configurations. The parent compound is LaFeO3 perovskite structure. It is Pnma symmetry
Lattice (Primitive)
a 5.60 Å, b 5.66 Å, c 7.94 Å, α 90.00 º, β 90.00 º, ɣ 90.00 ºWyckoff Element x y z
4b Fe 1/2 0 1/2
4c La 0.009381 0.961741 3/4
4c O 0.08612 0.478265 1/4
8d O 0.712594 0.287975 0.453725so all the atom substitutions of Ni, Co, Cu and Mn will go into that 4b Fe site. From the analysis, it has 4 symmetry equivalent sites:
5 T 26 FE 0.000000000000E+00 0.000000000000E+00 -5.000000000000E-01 6 F 26 FE -5.000000000000E-01 0.000000000000E+00 -3.095458132887E-17 7 F 26 FE -5.000000000000E-01 -5.000000000000E-01 2.220446049250E-16 8 F 26 FE 0.000000000000E+00 -5.000000000000E-01 -5.000000000000E-01I know the Fe composition, it is 20 %, the rest are Ni, Co, Cu and Mn, all at the same 20 % compositions - this simplifies the problem. But then I have 4 equivalent Fe sites and 5 elements to fit. Am I correct that I need to construct a supercell here? Ultimately, I would like this module to return me possible combinations of all 20% five elements, but not sure what size of supercell to create
see the attached test structure input.out INPUT.d12
thank you
-
Is there a software to create DOS/BAND input?I admit I am not very good conceptualizing these symetry unique points on Brillouin zone or expressing them in .d3 file as a path. I have two structures below that I fully optimized and calculated vibrational frequencies. Now my task is DOS/BAND. Is there a GUI that would help me read the optimized structures and prepare .d3 file? Or can I use my optimization file somehow to extract those unique points?
SPACE GROUP (CENTROSYMMETRIC) : P 21/N
LATTICE PARAMETERS (ANGSTROMS AND DEGREES) - CONVENTIONAL CELL
A B C ALPHA BETA GAMMA
7.40400 21.53400 5.65700 90.00000 91.05000 90.00000and
SPACE GROUP (CENTROSYMMETRIC) : P -1
LATTICE PARAMETERS (ANGSTROMS AND DEGREES) - CONVENTIONAL CELL
A B C ALPHA BETA GAMMA
10.70800 10.11400 6.41000 91.30000 88.30000 91.00000 -
lots of negative energy DE in crystal with good gradientsDear all, I optimized a crystal, got good gradients, but numerical displacements are already suspect as they generate lots of negative energies, it's a big system, I am still running it but decided to preemptively ask why this can be the case?
SYMMETRY ALLOWED FORCES (COORDINATE, FORCE)
1 -7.1019812E-07 2 2.6847537E-07 3 1.7714543E-06 4 1.5380909E-06 5 -8.5713887E-07 6 -7.6313741E-07 7 -1.3777999E-06 8 -4.2292032E-07 9 3.7330006E-07 10 -1.0186152E-07 11 7.8436370E-08 12 -1.0666144E-0713 7.6024231E-07 14 1.2549944E-07 15 -3.8114243E-07 16 8.3483245E-07
17 1.2719080E-06 18 1.3589256E-07 19 -1.2770755E-06 20 -1.7657611E-06
21 -6.7831105E-07 22 -2.8553633E-06 23 -2.5801301E-06 24 1.6190965E-06
25 2.7986374E-07 26 1.6445538E-06 27 1.5791380E-06 28 3.4913230E-06
29 7.5492284E-07 30 -3.2944974E-06 31 -2.9919538E-06 32 1.2296182E-06
33 4.5466078E-07 34 -1.8564773E-07 35 9.3158783E-07 36 -1.4184621E-07
37 2.8763484E-06 38 -1.7874331E-06 39 -3.3548919E-07 40 -1.0246636E-06
41 7.3025290E-07 42 -7.4914105E-07 43 -1.5527795E-07 44 -5.9189571E-07
45 1.1402401E-07 46 1.1445077E-07 47 5.0641463E-07 48 2.6720325E-07
49 3.9382990E-07 50 1.9265418E-07 51 -5.0025563E-07 52 -1.4722329E-07
53 -3.9032570E-07 54 -1.8660950E-07 55 -3.7294087E-08 56 2.7903845E-07
57 -1.1262719E-07 58 -1.4056720E-06 59 -7.2814440E-07 60 -4.2360016E-07
61 9.6760468E-07 62 -1.1476558E-06 63 5.7400637E-08 64 8.2334616E-07
65 1.7677814E-06 66 1.1935613E-06 67 -5.3996977E-07 68 5.3577787E-07
69 -5.5566146E-07 70 2.5231789E-07 71 -6.3487986E-07 72 7.2284427E-07
73 -8.4328694E-08 74 -5.8224781E-07 75 -1.6946265E-07 76 1.0209263E-06
77 2.9716506E-07 78 2.3838462E-07 79 1.7955521E-06 80 4.0804899E-07
81 1.2100282E-07 82 -1.5099145E-06 83 2.9006726E-07 84 -2.2790446E-07
85 1.4325087E-06 86 -6.8494225E-07 87 -3.3477344E-07 88 -2.2149944E-07
89 1.3328321E-07 90 -1.8330282E-08ATOM MAX ABS(DGRAD) TOTAL ENERGY (AU) N.CYC DE SYM
CENTRAL POINT -5.463640944184E+03 0 0.0000E+00 8
1 O DX 6.5777E-04 -5.463640944692E+03 11 -5.0781E-07 1
1 O DY 8.9722E-04 -5.463640943824E+03 12 3.6002E-07 1
1 O DZ 2.7715E-03 -5.463640938222E+03 13 5.9626E-06 1
9 O DX 1.4159E-03 -5.463640942796E+03 11 1.3886E-06 1
9 O DY 2.4328E-03 -5.463640939470E+03 13 4.7142E-06 1
9 O DZ 7.5341E-04 -5.463640944273E+03 10 -8.8837E-08 1
17 N DX 3.3298E-03 -5.463640936786E+03 9 7.3979E-06 1
17 N DY 2.6834E-03 -5.463640938932E+03 10 5.2525E-06 1
17 N DZ 2.2061E-03 -5.463640940002E+03 10 4.1826E-06 1
25 H DX 5.8163E-04 -5.463640944625E+03 10 -4.4035E-07 1
25 H DY 9.7068E-04 -5.463640943311E+03 9 8.7315E-07 1
25 H DZ 1.3265E-03 -5.463640942408E+03 11 1.7757E-06 1
33 H DX 1.9126E-03 -5.463640940840E+03 12 3.3445E-06 1
33 H DY 9.0051E-04 -5.463640944155E+03 9 2.8878E-08 1
33 H DZ 6.4300E-04 -5.463640945217E+03 9 -1.0328E-06 1
41 N DX 3.4011E-03 -5.463640936782E+03 10 7.4018E-06 1
41 N DY 2.7133E-03 -5.463640938661E+03 12 5.5236E-06 1
41 N DZ 2.8802E-03 -5.463640937601E+03 11 6.5830E-06 1
49 H DX 1.5489E-03 -5.463640941848E+03 10 2.3365E-06 1
49 H DY 3.3487E-04 -5.463640945255E+03 9 -1.0709E-06 1
49 H DZ 8.9868E-04 -5.463640943999E+03 9 1.8543E-07 1
57 N DX 2.4112E-03 -5.463640939472E+03 9 4.7126E-06 1
57 N DY 3.5207E-03 -5.463640936173E+03 10 8.0110E-06 1
57 N DZ 3.2454E-03 -5.463640936926E+03 12 7.2582E-06 1
65 H DX 4.2444E-04 -5.463640945298E+03 10 -1.1135E-06 1
65 H DY 1.4990E-03 -5.463640942117E+03 10 2.0672E-06 1
65 H DZ 9.8569E-04 -5.463640943558E+03 7 6.2581E-07 1
73 H DX 8.8996E-04 -5.463640943796E+03 11 3.8818E-07 1
73 H DY 4.0635E-04 -5.463640945249E+03 9 -1.0646E-06 1
73 H DZ 1.5526E-03 -5.463640941922E+03 11 2.2623E-06 1
81 N DX 3.0237E-03 -5.463640937800E+03 12 6.3840E-06 1
81 N DY 3.1621E-03 -5.463640937605E+03 12 6.5795E-06 1
81 N DZ 3.3672E-03 -5.463640936782E+03 11 7.4017E-06 1
89 H DX 3.5908E-04 -5.463640945209E+03 8 -1.0249E-06 1
89 H DY 1.0305E-03 -5.463640943313E+03 13 8.7074E-07 1
89 H DZ 1.3452E-03 -5.463640942571E+03 10 1.6132E-06 1
97 H DX 1.1682E-03 -5.463640943082E+03 10 1.1022E-06 1
97 H DY 1.4461E-03 -5.463640941923E+03 12 2.2616E-06 1output.out attached
